The function will take a function that has an occurrence dataset as an argument, and reruns it iteratively on the subsets of the dataset.
Usage
subsample(
x,
q,
tax = NULL,
bin = NULL,
FUN = divDyn,
coll = NULL,
iter = 50,
type = "cr",
keep = NULL,
rem = NULL,
duplicates = TRUE,
output = "arit",
useFailed = FALSE,
FUN.args = NULL,
na.rm = FALSE,
counter = TRUE,
...
)Arguments
- x
(
data.frame): Occurrence dataset, withbin,taxandcollas column names.- q
(
numeric): Subsampling level argument (mandatory). Depends on the subsampling function, it is the number of occurrences for"cr", and the number of desired occurrences to the power ofxexpfor O^x^W. It is also the quorum of the SQS method.- tax
(
character): The name of the taxon variable.- bin
(
character): The name of the subsetting variable (has to be integer). For time series, this is the time-slice variable. Rows withNAentries in this column will be omitted.- FUN
(
function): The function to be iteratively executed on the results of the subsampling trials. If set toNULL, no function will be executed, and the subsampled datasets will be returned as alist. By default set to thedivDynfunction. The function must have an argument calledx, that represents the dataset resulting from a subsampling trial (or the entire dataset). Arguments of thesubsamplefunction call will be searched for potential arguments of this function, which means that already provided variables (e.g.binandtax) will also be used. You can also provide additional arguments (similarly to theapplyiterator). Functions that allow arguments to pass through (that have argument '...') are not allowed, as well as functions that have the same arguments assubsamplebut would require different values.- coll
(
character): The variable name of the collection identifiers.- iter
(
numeric): The number of iterations to be executed.- type
(
character): The type of subsampling to be implemented. By default this is classical rarefaction ("cr"). ("oxw") stands for occurrence weighted by-list subsampling. If set to ("sqs"), the program will execute the shareholder quorum subsampling algorithm as it was suggested by Alroy (2010). Setting the argument to"none"will invoke no subsamling, but the applied function will be iterated on the trials, nevertheless.- keep
(
numeric): The bins which will not be subsampled but will be added to the subsampling trials. If the number of occurrences does not reach the subsampling quota, by default it will not be represented in the subsampling trials. You can force their inclusion with thekeepargument separetely (for all, see theuseFailedargument).- rem
(
numeric): The bins, which will be removed from the dataset before the subsampling trials.- duplicates
(
logical): Toggles whether multiple entries from the same taxon ("tax") and collection ("coll") variables should be omitted. Useful for omitting occurrences of multiple species-level occurrences of the same genus. By default these are allowed through analyses (duplicates=TRUE), setting this toFALSEwill require you to provide a collection variable. (coll)- output
(
character): If the function output are vectors or matrices, the"arit"and"geom"values will trigger simple averaging with arithmetic or geometric means. If the function output of a single trial is again avectoror amatrix, setting the output to"dist"will return the calculated results of every trial, organized in alistof independent variables (e.g. if the function output is value, the return will contain a singlevector, if it is avector, the output will be a list ofvectors, if the function output is adata.frame, the output will be alistofmatrixclass objects). Ifoutput="list", the structure of the original function output will be retained, and the results of the individual trials will be concatenated to alist.- useFailed
(
logical): If the bin does not reach the subsampling quota, should the bin be used?- FUN.args
(
list): Arguments passed to the applied functionFUNbut not used by the subsampling wrapper. Normally, the arguments ofFUNcan be added to the call ofsubsample, but in case you want to use different values for the same argument, then the arguments added here will be used for the call ofFUN. For instance, if you want to callsubsamplewithbin=NULL, but want to runFUN=divDynwith a validbincolumn then you can add the column name here, e.g.FUN.args=list(bin="stg").- na.rm
(
logical): The function call includes more column names that might contain missing values. If this flag is set toTRUE, all rows will be dropped that have missig values in the specificed columns. This might lead to the exclusion of some data you do not want to exclude.- counter
(
logical): Should the loop counting be visible?- ...
arguments passed to
FUNand the type-specific subsampling functions:subtrialCR,subtrialOXW,subtrialSQS
Details
The subsample function implements the iterative framework of the sampling standardization procedure.
The function 1. takes the dataset x, 2. runs function FUN on the dataset and creates a container for results of trials
3. runs one of the subsampling trial functions (e.g. subtrialCR) to get a subsampled 'trial dataset'
4. runs FUN on the trial dataset and
5. averages the results of the trials for a simple output of step 4. such as vectors, matrices and data.frames. For averaging, the vectors and matrices have to have the same output dimensions in the subsampling, as in the original object. For data.frames, the bin-specific information have to be in rows and the bin numbers have to be given in a variable bin in the output of FUN.
For a detailed treatment on what the function does, please see the vignette ('Handout to the R package 'divDyn' v0.5.0 for diversity dynamics from fossil occurrence data'). Currently the Classical Rarefaction ("cr", Raup, 1975), the occurrence weighted by-list subsampling ("oxw", Alroy et al., 2001) and the Shareholder Quorum Subsampling methods are implemented ("sqs", Alroy, 2010).
References:
Alroy, J., Marshall, C. R., Bambach, R. K., Bezusko, K., Foote, M., Fürsich, F. T., … Webber, A. (2001). Effects of sampling standardization on estimates of Phanerozoic marine diversification. Proceedings of the National Academy of Science, 98(11), 6261-6266.
Alroy, J. (2010). The Shifting Balance of Diversity Among Major Marine Animal Groups. Science, 329, 1191-1194. https://doi.org/10.1126/science.1189910
Raup, D. M. (1975). Taxonomic Diversity Estimation Using Rarefaction. Paleobiology, 1, 333-342. https: //doi.org/10.2307/2400135
Examples
data(corals)
data(stages)
# Example 1-calculate metrics of diversity dynamics
dd <- divDyn(corals, tax="genus", bin="stg")
rarefDD<-subsample(corals,iter=30, q=50,
tax="genus", bin="stg", output="dist", keep=95)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
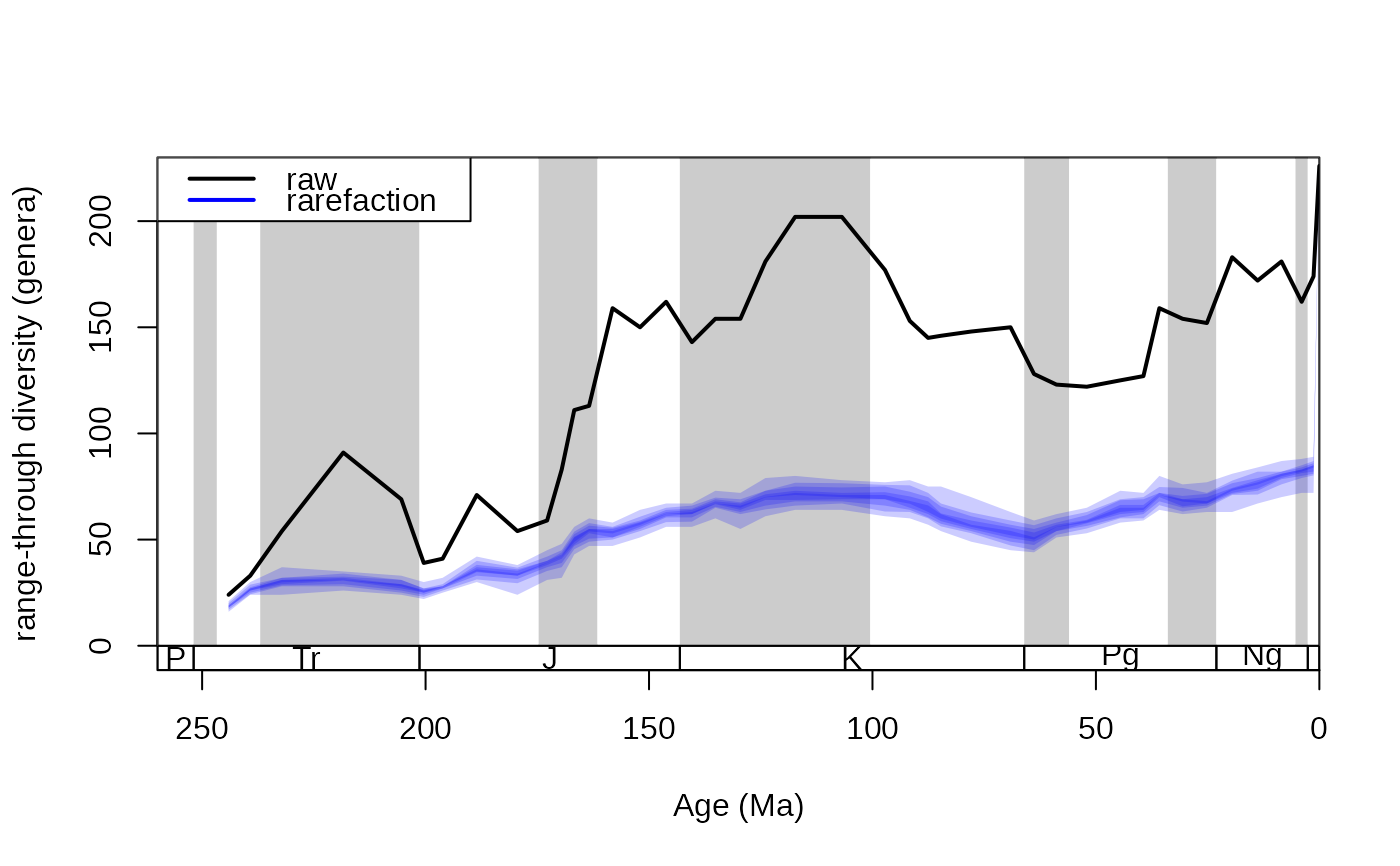
# plotting
tsplot(stages, shading="series", boxes="sys", xlim=c(260,0),
ylab="range-through diversity (genera)", ylim=c(0,230))
lines(stages$mid, dd$divRT, lwd=2)
shades(stages$mid, rarefDD$divRT, col="blue")
legend("topleft", legend=c("raw","rarefaction"),
col=c("black", "blue"), lwd=c(2,2), bg="white")
 # \donttest{
# Example 2-SIB diversity
# draft a simple function to calculate SIB diversity
sib<-function(x, bin, tax){
calc<-tapply(INDEX=x[,bin], X=x[,tax], function(y){
length(levels(factor(y)))
})
return(calc[as.character(stages$stg)])
}
sibDiv<-sib(corals, bin="stg", tax="genus")
# calculate it with subsampling
rarefSIB<-subsample(corals,iter=25, q=50,
tax="genus", bin="stg", output="arit", keep=95, FUN=sib)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
rarefDD<-subsample(corals,iter=25, q=50,
tax="genus", bin="stg", output="arit", keep=95)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
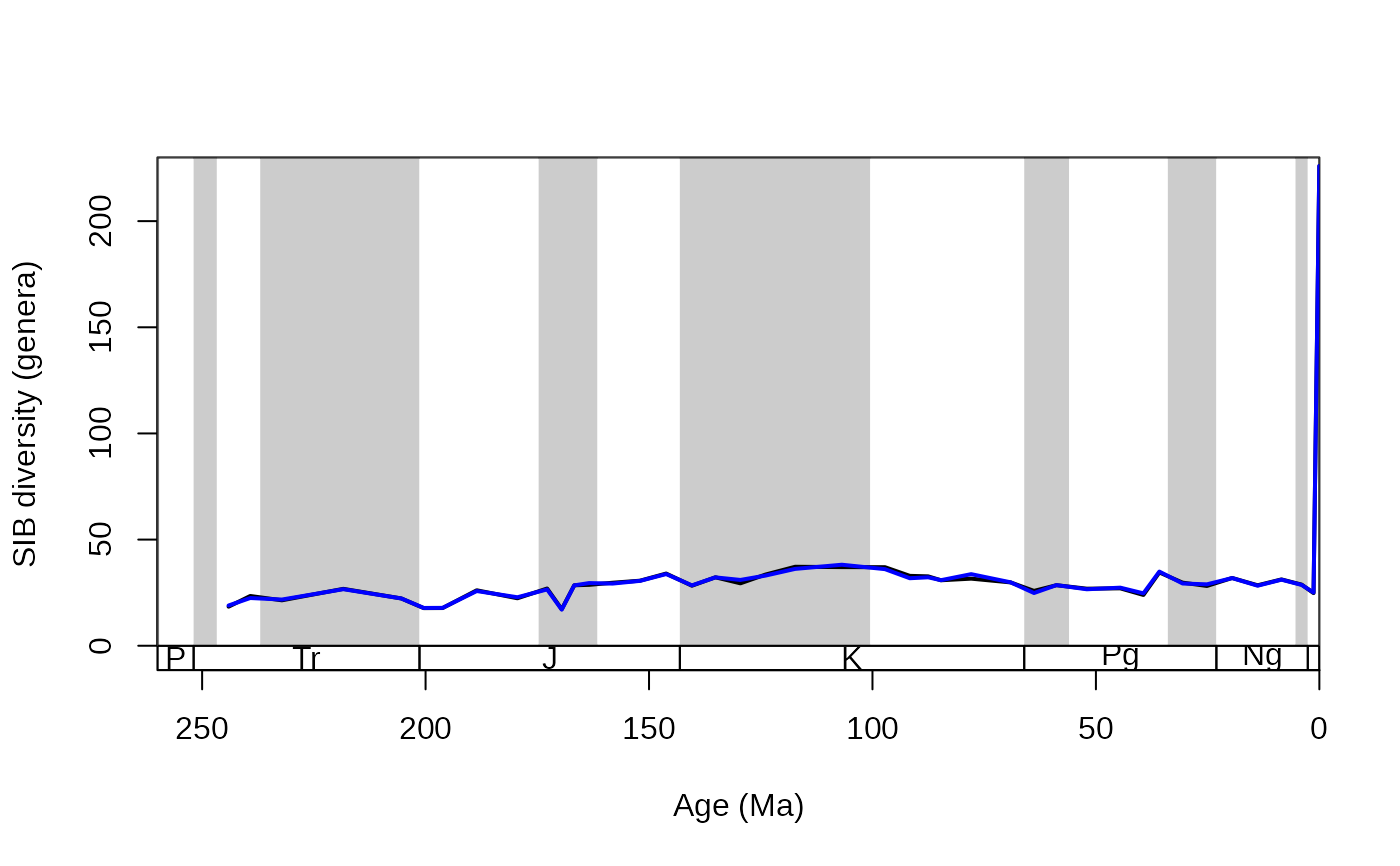
# plot
tsplot(stages, shading="series", boxes="sys", xlim=c(260,0),
ylab="SIB diversity (genera)", ylim=c(0,230))
lines(stages$mid, rarefDD$divSIB, lwd=2, col="black")
lines(stages$mid, rarefSIB, lwd=2, col="blue")
# \donttest{
# Example 2-SIB diversity
# draft a simple function to calculate SIB diversity
sib<-function(x, bin, tax){
calc<-tapply(INDEX=x[,bin], X=x[,tax], function(y){
length(levels(factor(y)))
})
return(calc[as.character(stages$stg)])
}
sibDiv<-sib(corals, bin="stg", tax="genus")
# calculate it with subsampling
rarefSIB<-subsample(corals,iter=25, q=50,
tax="genus", bin="stg", output="arit", keep=95, FUN=sib)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
rarefDD<-subsample(corals,iter=25, q=50,
tax="genus", bin="stg", output="arit", keep=95)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
# plot
tsplot(stages, shading="series", boxes="sys", xlim=c(260,0),
ylab="SIB diversity (genera)", ylim=c(0,230))
lines(stages$mid, rarefDD$divSIB, lwd=2, col="black")
lines(stages$mid, rarefSIB, lwd=2, col="blue")
 # Example 3 - different subsampling types with default function (divDyn)
# compare different subsampling types
# classical rarefaction
cr<-subsample(corals,iter=25, q=20,tax="genus", bin="stg", output="dist", keep=95)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
# by-list subsampling (unweighted) - 3 collections
UW<-subsample(corals,iter=25, q=3,tax="genus", bin="stg", coll="collection_no",
output="dist", keep=95, type="oxw", xexp=0)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
# occurrence weighted by list subsampling
OW<-subsample(corals,iter=25, q=20,tax="genus", bin="stg", coll="collection_no",
output="dist", keep=95, type="oxw", xexp=1)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
SQS<-subsample(corals,iter=25, q=0.4,tax="genus", bin="stg", output="dist", keep=95, type="sqs")
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
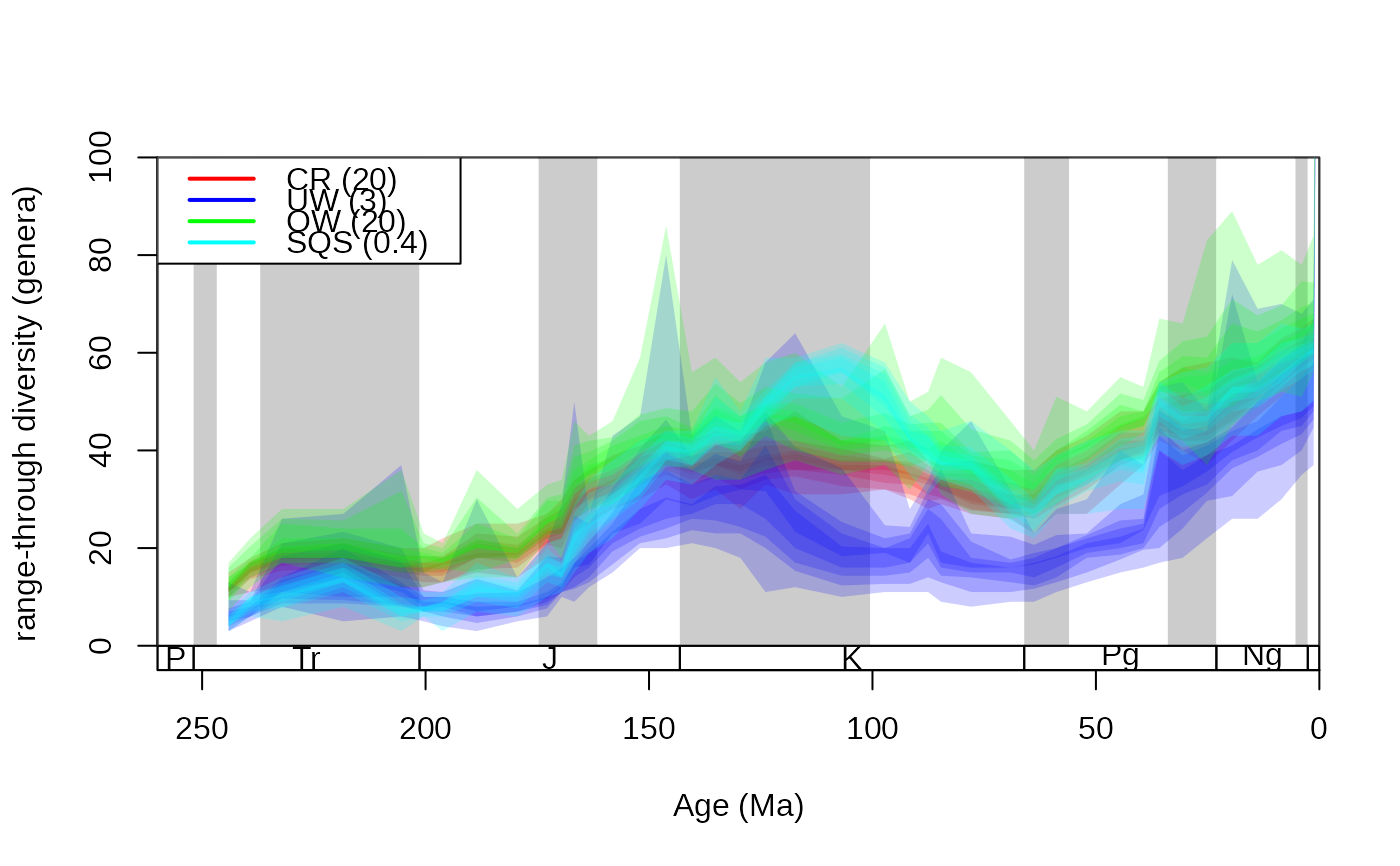
# plot
tsplot(stages, shading="series", boxes="sys", xlim=c(260,0),
ylab="range-through diversity (genera)", ylim=c(0,100))
shades(stages$mid, cr$divRT, col="red")
shades(stages$mid, UW$divRT, col="blue")
shades(stages$mid, OW$divRT, col="green")
shades(stages$mid, SQS$divRT, col="cyan")
legend("topleft", bg="white", legend=c("CR (20)", "UW (3)", "OW (20)", "SQS (0.4)"),
col=c("red", "blue", "green", "cyan"), lty=c(1,1,1,1), lwd=c(2,2,2,2))
# Example 3 - different subsampling types with default function (divDyn)
# compare different subsampling types
# classical rarefaction
cr<-subsample(corals,iter=25, q=20,tax="genus", bin="stg", output="dist", keep=95)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
# by-list subsampling (unweighted) - 3 collections
UW<-subsample(corals,iter=25, q=3,tax="genus", bin="stg", coll="collection_no",
output="dist", keep=95, type="oxw", xexp=0)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
# occurrence weighted by list subsampling
OW<-subsample(corals,iter=25, q=20,tax="genus", bin="stg", coll="collection_no",
output="dist", keep=95, type="oxw", xexp=1)
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
SQS<-subsample(corals,iter=25, q=0.4,tax="genus", bin="stg", output="dist", keep=95, type="sqs")
#> 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
# plot
tsplot(stages, shading="series", boxes="sys", xlim=c(260,0),
ylab="range-through diversity (genera)", ylim=c(0,100))
shades(stages$mid, cr$divRT, col="red")
shades(stages$mid, UW$divRT, col="blue")
shades(stages$mid, OW$divRT, col="green")
shades(stages$mid, SQS$divRT, col="cyan")
legend("topleft", bg="white", legend=c("CR (20)", "UW (3)", "OW (20)", "SQS (0.4)"),
col=c("red", "blue", "green", "cyan"), lty=c(1,1,1,1), lwd=c(2,2,2,2))
 # }
# }
