Function to quickly omit single-collection and single-reference taxa.
Arguments
- x
(data.frame)Occurrence dataset, withbin,taxandcollas column names.- om
(character)The type of omission."coll"omits occurrences of taxa that occurr only in one collection."ref"omits occurrences of taxa that were described only in one reference."binref"will omit the set of single reference taxa that were described by more than one references, but appear in only one reference in a time bin.- tax
(character)The name of the taxon variable.- bin
(character)The name of the bin variable (has to benumericfor the function to run). For time series, this is the time slice variable.- coll
(character)The variable name of the collection identifiers.- ref
(character)The variable name of the reference identifiers (optional).- filterNA
(logical)Additional entries can be added to influence the dataset that might not have reference or collection information (NAentries). These occurrences are treated as single-collection or single-reference taxa if thena.rmargument is set toFALSE(default). Setting this argument toTRUEwill keep these entries. (see example)
Details
The function returns a logical vector, with a value for each row. TRUE values indicate rows to be omitted, FALSE values indicate rows to be kept. The function is embedded in the divDyn function, but can be called independently.
Examples
# omit single-reference taxa
data(corals)
data(stages)
toOmit <- omit(corals, bin="stg", tax="genus", om="ref", ref="reference_no")
x <- corals[!toOmit,]
# within divDyn
# plotting
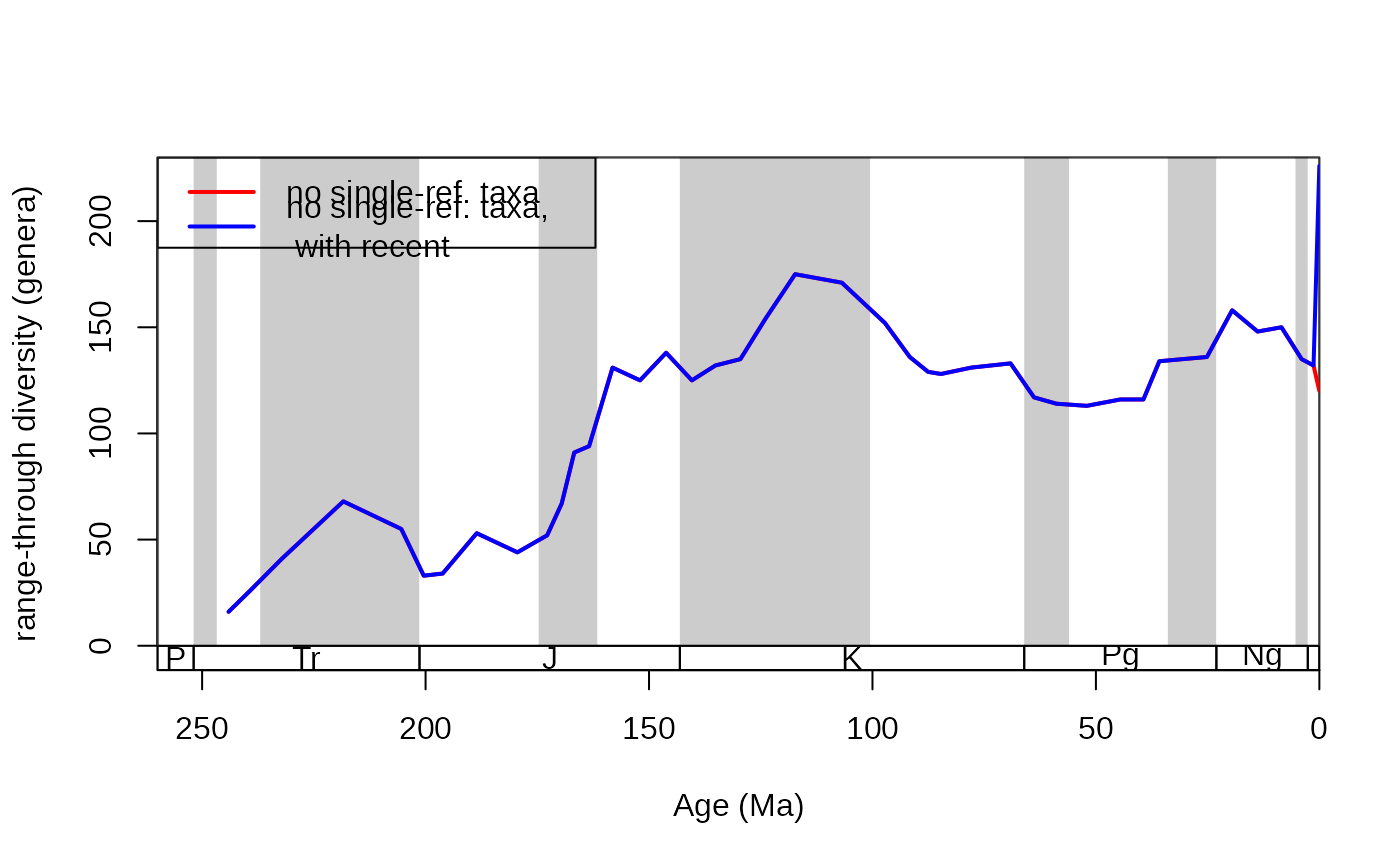
tsplot(stages, shading="series", boxes="sys", xlim=c(260,0),
ylab="range-through diversity (genera)", ylim=c(0,230))
# multiple ref/slice required
ddNoSing <- divDyn(corals, tax="genus", bin="stg", om="binref", ref="reference_no")
lines(stages$mid, ddNoSing$divRT, lwd=2, col="red")
# with the recent included (NA reference value)
ddNoSingRec <- divDyn(corals, tax="genus", bin="stg",
om="binref", filterNA=TRUE,ref="reference_no")
lines(stages$mid, ddNoSingRec$divRT, lwd=2, col="blue")
# legend
legend("topleft", legend=c("no single-ref. taxa",
"no single-ref. taxa,\n with recent"),
col=c("red", "blue"), lwd=c(2,2))